On this page
- Introduction to Cytogenetics
- Blood Culture and Karyotyping
- Referral Categories:
- Preferred volumes of blood
- Urgent Samples
- Reporting of results
- Postnatal SNP Array
- Prenatal Diagnosis
- Sample Requirements
- Sample Processing
- Reporting of Results
- Rapid Prenatal Diagnosis
- Tissue Samples
- Testing Pathways
- Reporting
- Taking and Sending Samples
- Sample Acceptance and Consent
- Ordering Medium
- How to order
The North East and Yorkshire Genomic Laboratory Hub (Central Laboratory) (NEYGLH) provides testing for a large number of genetic disorders, covering both cytogenetic and molecular genetic analysis. Please see individual sections for further information.
Introduction to Cytogenetics
Cytogenetics is the study of chromosomes and chromosomal abnormalities. The normal number of chromosomes in a human cell is 46. Chromosomes are made from DNA, and contain all the genes present in a cell. Each gene is located at a specific site on each chromosome. There are many clinical syndromes that have been associated with particular chromosomal abnormalities. The image below shows a normal male chromosome complement.
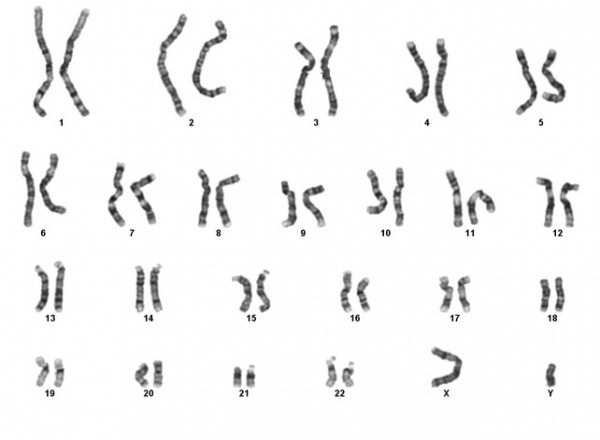
Chromosomal abnormalities usually involve large amounts of DNA and therefore tend to involve large numbers of genes. For this reason, disorders associated with chromosomal abnormalities are usually very serious and can involve multiple organs. Often these abnormalities are severe enough to be fatal, either in utero or in early life. Each normal human cell contains 23 pairs of chromosomes. 22 of these pairs are identical, the other constitutes the sex chromosomes (one X and one Y in a male and two Xs in a female).
The largest chromosome in the human complement is numbered ‘1’ and the smallest ’22’. Modern techniques within the laboratory allow the chromosomes to be identified from one another by distinguishing bands on the chromosomes.
The work of the department can be broken down into the following main areas.
Blood culture and karyotyping
This category comprises the major part of the workload. Urgent samples include those from newborn babies and pregnant women and their partners. Non-urgent referrals include patients with a history of infertility, individuals with impaired physical or sexual development, patients with a family history of a known chromosome abnormality, follow-up from prenatal diagnosis (confirmation of abnormal prenatal results only), screening of patients and donors for IVF and patients with a suspected chromosome fragility syndrome.
SNP array
SNP array is provided as a first line for test for patients for paediatric patients with learning difficulties, behavioural problems, developmental delay, autism, seizures and/or dysmorphism. Newborns may also be tested with congenital abnormalities at the clinicians request. Professional judgment will be used to determine which patients are tested in line with current laboratory policy.
Prenatal Diagnosis
Prenatal diagnosis is carried out by one of three methods – chorionic villus biopsy, amniocentesis or fetal blood sampling. Indications for prenatal diagnosis include advanced maternal age, problems detected on ultrasound scan, risks identified through measuring biochemical markers, and the presence of a family history of a chromosome abnormality. The main abnormality detected is Down Syndrome.
Solid Tissues
Since up to 50% of all spontaneous abortions may have a chromosome abnormality, products of conception after a pregnancy loss may be referred for chromosome analysis. Skin biopsies from living persons are also occasionally received either for cytogenetic analysis or for referral onwards for biochemical testing or DNA studies.
Blood Culture and Karyotyping
Referral Categories:
The main referral categories include:
- Abnormal newborns
- Investigation of infertility
- Parents of children with a known chromosome abnormality
- Family follow-up studies
- Screening of patients and donors for IVF
Referral for:
Test Requirements:
Sample Tubes
Blood samples for chromosome analysis should be collected in a lithium heparin tube (orange or green top), and mixed well to prevent clotting. It is important to ensure that the sample tube contains lithium heparin irrespective of the colour of the tube top.
Blood samples for SNP array should be collected in EDTA.
Preferred volumes of blood
- Adults 5ml
- Children 2 to 5ml
- Infants 1 to 2ml
More blood may be needed if chromosome breakage studies are required.
Sample Transport
Please see laboratory contact page for address details.
Cytogenetic analysis requires living cells. Please ensure that the sample reaches us as quickly as possible (within 48 hours). First class post is satisfactory for non-urgent samples.
Urgent samples should be sent by courier or taxi.
Samples should not be frozen, exposed to excess heat or stored in formalin.
If there is a delay in transit please store the sample at 4°C (in a refrigerator).
Please try to avoid sending samples at weekends or bank holidays as post is not delivered on these days.
When sending samples by post a secure container should be used to conform to current postal regulations, i.e. P650 and UN3373 applicable.
Urgent Samples
Cases treated as urgent are all new borns, cordocentesis specimens, pregnant couples including follow up of prenatal diagnosis, children awaiting surgery and others cases as discussed with the laboratory.
Reporting of results
Urgent cases
Usually within 10 days which is within the national guidelines. An aneuploidy screen can often be done overnight as a QF-PCR or Fluorescent in situ Hybridisation (FISH) test (please contact the laboratory).
Other cases
We attempt to report within the national guidelines which are within 42 calendar days. If results are required earlier please contact the lab.
Complex abnormalities, or abnormalities that are difficult to interpret, may require FISH or SNP array to resolve the karyotype, this may delay some results.
Results are sent to the referring clinician. Complex abnormal results are usually telephoned prior to the written report being sent and the interpretation and implication discussed. Abnormal results from urgent cases will be telephoned to the referring clinician.
In response to telephone enquiries, only normal results or those which confirm a previous finding are given to a clinician’s secretary or the clinic sister. All other results are only given to clinicians.
Postnatal SNP Array
Alternative testing for samples suboptimal for SNP Array
A technique, referred to as CNVseq (Copy Number Variant detection by Sequencing), was introduced into the laboratory in January 2014 to complement array testing. The method uses a Next Generation Sequencing approach to look for copy number variation in the genome in a similar way to SNP array. The technique is used for samples which are considered to be suboptimal for array due to poor quality/low amount of DNA input. These samples are tested by CNVseq instead of array, with the same guidelines for analysis and reporting being applied. Due to the nature of the test, the resolution obtained per case will vary; this will be stated in the report. Please note that the test is currently performed only once every month. As a result, the turnaround times may be slightly higher compared to SNP array.
Fragile X testing
Referrals for patients with a genetically confirmed family history of Fragile X (clinical information of affected family member must be provided) will be tested on request.
Since 1st August 2015, all other samples have array testing performed as a first line test as it has a higher detection rate of causative abnormalities than the Fragile X test. It cannot, however, detect the Fragile X mutation. If following a normal array result Fragile X testing is still required, a newly completed referral form must be sent to the laboratory in order for testing to be initiated.
Incidental findings
As SNP array is a whole genome screen there may be occasional unexpected incidental findings which are medically important but unrelated to the specific reasons for referral. It is important to explain to the family that the test is a very detailed look at the whole of the genome and occasionally unexpected incidental findings are detected.
Clinical advice
Clinical advice is available from the Department of Clinical Genetics, Chapel Allerton Hospital, Leeds. The team are happy to receive referral letters for patients that have had abnormal array results and/or where there are clinical implications for other family members.
The following test is accredited:
SNP array testing
This test is now accredited to ISO15189 by UKAS.
Prenatal Diagnosis
Reasons for Referral
Increased risk of chromosome abnormality, including:
- Abnormal ultrasound scan
- Increased risk following screening, (biochemical or ultrasound)
- Parent carries a chromosome rearrangement
- Previous child with a chromosome abnormality
Samples are also accepted from patients whose pregnancy is at risk of a non-chromosomal genetic or metabolic disorder. These patients should usually be referred through the Dept of Clinical Genetics at Chapel Allerton Hospital, Leeds and as much advance notice as possible must always be given to the laboratory.
Please Note:
Please state clearly on the referral card if a test other than standard Cytogenetics is required, for example testing for cystic fibrosis where there is a scan finding of echogenic bowel.
If any of the original sample or cultured cells are to be forwarded to another laboratory, please state this on the referral card, giving details of where the sample is to be sent, what test is required, and what the transport requirements are. Costs of tests carried out elsewhere as well as transport costs are usually met by the referring clinician.
If possible patients with a family history of a chromosome or other relevant genetic abnormality should be screened by the investigation of parental blood samples before prenatal diagnosis is undertaken.
Sample Transport
Please see contact page for laboratory address and contact details.
Cytogenetic analysis requires living cells. Please ensure that the sample reaches us as quickly as possible (preferably within 24 hours). First class post is satisfactory.
Samples should not be frozen or exposed to excess heat. If there is a delay in transit please store the sample at 4C (in a refrigerator).
Please try to avoid sending samples at weekends or bank holidays.
When sending samples by post a secure container should be used to conform to current postal regulations, i.e. P650 and UN3373 applicable.
Sample Requirements
Amniotic Fluids
10-20mls of fluid (15-20mls if SNP array testing is required – see later for further details) in a sterile, leak proof, plain (no anticoagulant), plastic universal container labelled with at least two unique patient identifiers (name, NHS number, date of birth, hospital number)
A completed constitutional testing referral form must accompany all samples. It is important that all information requested on the card is supplied by the referring hospital; failure to provide relevant information may result in delays in reporting results.
Samples must reach the laboratory by 3pm for a rapid (QFPCR) result to be available the next working day.
Chorionic Villus Samples (CVS)
CVS should be received on the same day taken and preferably no later than 4pm.
CVS must be sent in transport medium, details of which can be supplied by the laboratory.
Cytogenetic studies are performed on CVS with an optimal size of 10mg or more. For other investigations such as DNA or biochemical investigations, 20mg or more are preferred.
Where samples have been referred primarily for SNP array, DNA or biochemical testing, cytogenetic studies will only be performed if there are sufficient villi.
A completed constitutional testing referral form must accompany all samples as for amniotic fluids, above.
Fetal Blood Samples
Fetal blood samples should be collected in to 2ml paediatric lithium heparin tubes.
0.5 – 1.0ml is usually sufficient.
Samples should be accompanied by a completed cytogenetics referral form, as above.
Please state that the sample is fetal blood and whether or not the pregnancy is ongoing.
Maternal blood samples
A maternal blood sample (2 to 3ml in EDTA) should accompany all prenatal invasive samples to help with the interpretation of rapid results.
For requirements for parental karyotyping, please refer to separate user guide for postnatal samples.
Sample Processing
Amniotic fluid and chorionic villus samples will be tested using QFPCR for 13,18,21, X&Y. Samples will only be fully karyotyped in the event of an abnormal result.
Rapid FISH testing may be undertaken in addition to QFPCR for fetal heart defects detected on the 20 week anomoly scan.
Direct karyotype from chorionic villus material will be performed in cases which require a rapid full karyotype e.g. family history of a balanced chromosome rearrangement.
Following a normal QFPCR result, any pregnancy with an increased NT (>3.5 mm) or an abnormality detected on ultrasound scan will be eligible for SNP array testing.
Reporting of Results
Results obtained from QFPCR testing (of both CVS and amniotic fluids) and direct CVS preparations are usually available the following day as long as the sample was received early enough for processing. Cytogenetic results from direct CVS preparations are provisional, a final written report is only issued once the long term cultures have been analysed.
SNP array results will be available within 14 days unless DNA needs to be extracted from cultures which will delay the reporting time. The clinician will be contacted in the event of this happening to advise of the delay.
Fetal blood karyotype results are usually reported to the referring clinician within 4 to 5 working days following receipt of the sample. A provisional result may be available in 48 hours.
Amniotic fluid and CVS (full karyotype): The reporting time is dependent upon rate of cell growth in culture, but generally results are available in 2 weeks. These are sent to the referring clinician and will mostly be confirmations of abnormal QFPCR findings.
All rapid (QFPCR, direct CVS & FISH) and SNP array results will be emailed to the referring clinician and /or designated contact (eg screening co-ordinator).
Where abnormal results are complex or ambiguous, reports may also be sent to the Clinical Genetics Unit if this is considered appropriate.
Please note we no longer require a follow-up sample at delivery / termination for confirmation of abnormal prenatal results.
Maternal cell contamination is a limitation with amniotic fluid and chorionic villus culture. If unrecognised, this may lead to a misdiagnosis.
Prenatal SNP array Testing – ‘Information for the Clinician’
For further information please see the user guide.
Rapid Prenatal Diagnosis
What is QFPCR?
QFPCR stands for quantitative fluorescence polymerase chain reaction. Small sections (markers) of DNA from the sample are amplified, labelled with fluorescent tags and the amounts measured by electrophoresis.
What can it test for?
QFPCR is used to test for gene dosage – ie. the number of copies of a given gene present in a sample. The YNEGLH Central Lab uses QFPCR to test for aneuploidy of whole chromosomes – currently chromosomes 13, 18 and 21 – in amniotic fluid samples. It is possible to test for sex chromosomes by QFPCR but the Leeds lab does not currently offer this service.
What do the results look like?
The results are presented as a graph and ratios are calculated using a spreadsheet.
For each marker tested, a normal result is represented on the graph as two peaks of equal height and area (one peak for each chromosome present). So, for example, a normal pattern for the chromosome 13 marker D13S742 would look like this:
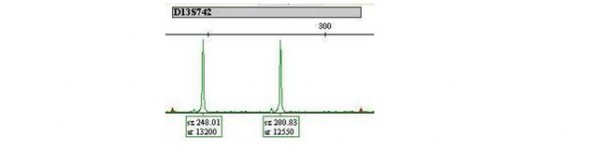
The name of the marker is given in the grey box above the peaks. The numbers underneath the peaks represent the length of the marker (sz) and the amount present (peak area, ar). So this sample has one D13S742 marker which is 248.01 bases long and another which is 280.83 bases long. The two copies are referred to as ‘alleles’ and there is one on each copy of chromosome 13, present in a ratio of 13200:12550 or approximately 1:1.
An example trace for a normal sample is shown below.
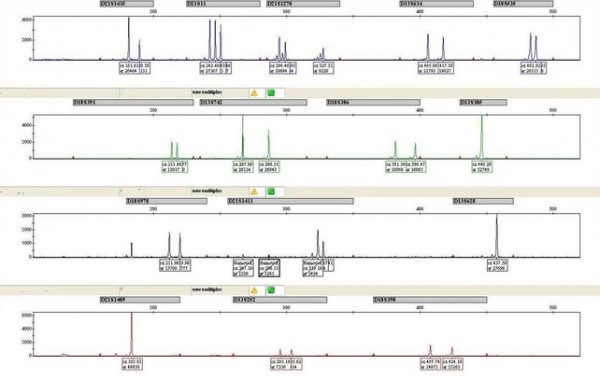
The four rows on the graph correspond to the four different fluorescent tags used to label the DNA – at least one marker per chromosome is present on each row, with five markers per chromosome in total.
In this example, the chromosome 21 marker D21S311 has only a single peak visible. This is because it has two alleles of the same length and the two peaks are superimposed. This means that a ratio cannot be calculated and the marker is uninformative.
If the sample is abnormal, a different pattern is seen on the trace:
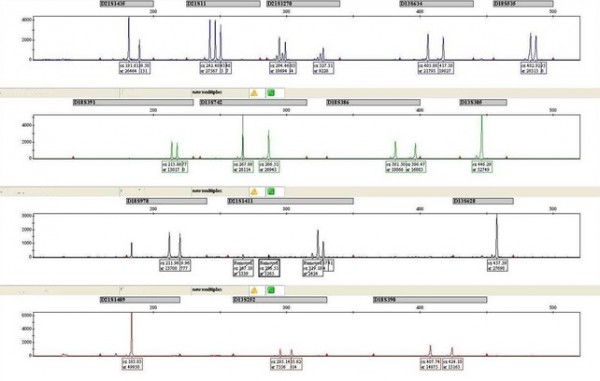
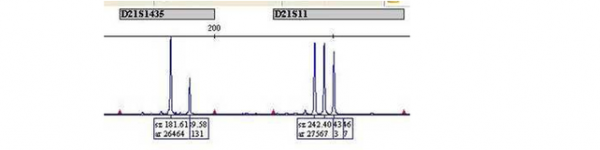
Markers D21S11 and D21S1411 show three peaks of equal area, indicating that there are three different alleles present in a 1:1:1 ratio. Marker D21S1435 shows only two peaks but the ratio of the peak area for the two alleles is 2:1. The 2:1 ratio also indicates the presence of three alleles, however two of the alleles are of the same length, and therefore superimposed as for marker D21S11 in the ‘normal’ example.
All of these results indicate the presence of three copies of chromosome 21, meaning that the foetus will have Down syndrome. Note that the chromosome 13 and chromosome 18 markers show a normal pattern.
Tissue Samples
Sample Requirements
Pregnancy losses
Ideally a sample of cord and/or placenta; post-mortem samples of skin, lung, cartilage etc (foetal samples). Products of conception may be sent if there is no alternative but please note that the results may be less accurate due to maternal cell contamination. If sending placental tissue or products of conception, please also send a small (2-3ml) sample of maternal blood in EDTA if possible; this will enable exclusion of maternal cell contamination and ensure that the result is reported appropriately.
Live patients (for mosaicism / enzymes / cell storage etc): A full thickness skin biopsy.
Please note that, due to the restrictions of the Human Tissue Act, whole foetuses cannot be accepted by the laboratory and will be returned to the referrer without processing. Products of conception containing recognisable foetal parts will be returned for burial / disposal once appropriate material has been processed.
Samples collected into formalin, those that are obviously infected, and samples which are excessively delayed in transit are unsuitable for cytogenetic studies and no attempt will be made to test such specimens.
A fully completed referral form must accompany all samples. Download a referral card from the top of this page.
Sample Transport
Whole foetuses are not accepted by the laboratory and, if sent, will be returned.
Samples should be collected into a sterile Universal container containing culture medium (Ham’s F10) and antibiotics. A supply of suitable transport medium can be obtained from the laboratory on request.
If no transport medium is available it is possible to send the tissue in sterile isotonic saline, or in a dry sterile vessel. These samples should be sent to the laboratory without delay.
Samples must not be frozen, exposed to excess heat or stored in formalin.
Samples should be addressed to: YNEGLH, Central lab. Please see laboratory contact details for address and contact details.
Some of the testing performed requires living cells. Please ensure that the sample reaches us as quickly as possible (within 48 hours). First class post is satisfactory.
If there is a delay in transit please store the sample at 4°C (in a refrigerator).
Please try to avoid sending samples at weekends or bank holidays.
When sending samples by post a secure container should be used to conform to current postal regulations, i.e. P650 and UN3373 applicable.
Testing Pathways
Obstetric samples (pregnancy loss / TOP / IUD / stillbirth):
From 1st October 2016, all obstetric samples will be tested by QFPCR (Elucigene QST*R-PLTM) for chromosomes 13/15/16/18/21/22/X/Y to identify those aneuploidies which are most commonly found in pregnancy losses.
From the 1st February 2021 a subset of those with normal QFPCR results will then be tested by SNP Array instead of CNV sequencing. SNP arrays can identify significant copy number imbalances including, but not limited to, whole chromosome aneuploidy, which are likely to have contributed to the pregnancy loss and/or fetal abnormality.
Samples which will receive the additional SNP array test are:
- Pregnancy losses at or before 12 weeks
- Pregnancy losses / TOP at any gestation where there is a fetal abnormality
- Where there is a history of 3 or more recurrent miscarriages
Audit data suggest that other referrals have a very low incidence of clinically significant copy number abnormalities detectable by SNP Array but not by QFPCR.
Please include as much information as possible on the referral form so that the sample is processed in the correct workstream.
Sudden infant deaths and skin biopsies from live patients:
These samples will continue to be cultured for karyotype analysis/ SNP Array / DNA storage / cell storage / metabolic testing as appropriate.
Reporting
All QFPCR results will be issued in the form of a written report with a 42 calendar day reporting time target. SNP Array results will be issued separately and may take up to a month longer since, due to batching requirements, we will only be performing a run every few weeks.
For SNP Array results the reporting criteria will be similar to those presently in use for prenatal SNP array, so that only those copy number variants which are thought to be directly relevant to the referral reason (i.e. likely to be a cause of the pregnancy loss or fetal anomaly) will be reported. Incidental findings and variants of uncertain significance will usually not be reported.
Results are sent by post to the referring clinician and / or pathologist. Complex abnormal results may be telephoned prior to the written report being sent and the interpretation and implication discussed.
In response to telephone enquiries, only normal results or those which confirm a previous finding can be given to a clinician’s secretary or the clinic sister. All other results are only given to clinicians or faxed reports are sent to designated contacts. Reports in pdf format can be emailed if a suitable secure nhs.net email address is supplied.
Please note
Although every effort is made to use suitable material for analysis, it is not always possible to avoid analysing maternal cells where the material used is of extra-embryonic origin (e.g. products of conception, placenta). Samples which are subject to this limitation will be reported with appropriate interpretative comments.
Taking and Sending Samples
Specimen Containers
For karyotype analysis we require 3mls of blood in a lithium heparin tube (green or orange lid).
For SNP array testing we require 3mls of blood in an EDTA tube (purple or pink lid).
Additional blood will be required if chromosome breakage studies are required (approx 2-3ml), dependent on the number and test types.
All specimen containers must be clearly labelled, by hand and in CAPITAL LETTERS with the following information:
- Forename
- Surname
- Date of Birth
- Hospital/NHS Number
- Ward/Location
- Date of collection
Sample Transportation
It is the responsibility of the person transporting the specimen to ensure that it arrives promptly and undamaged at its destination.
Samples should be sent in sealed plastic bags, with the request card protected from the sample.
The plastic bags should be placed in taped boxes containing appropriate padding and packing.
When sending samples by post a secure container should be used to conform to current postal regulations, i.e. P650 and UN3373 applicable. For more information please see Guide to the Packaging & Transportation of Biological Specimens by Road.
Packaging regulations (36kB pdf)
Please see the laboratory contact page for address and contact details.
Sample Acceptance and Consent
It is assumed that when a sample is sent to the laboratory, the clinician responsible for the care of the patient has obtained appropriate and valid consent for testing and storage so that the laboratory is not required to confirm and document such consent.
For more information on consent please see the guidelines from the Joint Committee on Medical Genetics:
Joint Committee on Medical GeneticsAcceptance of a sample and referral card from the referring clinician by the testing laboratory should be considered as an entry into an agreement between both parties.
It is the responsibility of the person (doctor, nurse or phlebotomist) taking the sample from the patient to ensure that the specimen container is correctly identified. Those sent in an incorrect container may not be accepted.
Incorrectly/unlabelled samples/referral cards may not be accepted by the laboratory.
The laboratory responsibility begins when the specimen arrives at YNEGLH Central Lab reception. The Leeds lab can only act upon the information provided and upon the accuracy of that information.
It is the responsibility of the referring clinician to clearly state on the referral card the test required. The referring clinician should also include any other information that may affect the interpretation of the result such as clinical features and family history.
The laboratory will inform the referring clinician as soon as practical if the sample is unsuitable for testing, the identifiers provided are insufficient or the test requested is unclear. It is the responsibility of the laboratory to report the result within the turnaround times indicated. Any factors leading to delay or quality of testing or should be communicated to the referring clinician as soon as practical.
The final decision on whether to accept a specimen will lie with the Head of the Laboratory.
Ordering Medium
We supply the following medium:
- Tissue and tumour transport medium
- CVS transport medium
- Marrow sample transport medium
Expiry date will be clearly labelled on the tube, but transport medium must be refrigerated on receipt.
Unused medium should be disposed of with clinical waste.
How to order
We need to know:
- The number of bottles
- Name and address to send to
- The type of medium requested
Telephone on 0113206 5419.